What is CellCap?
CellCap is a probabilistic generative model for explainable modeling of single-cell perturbation data. CellCap employs sparse dictionary learning in a latent space to deconstruct cell-state-specific perturbation responses into a set of transcriptional response programs. These programs are then utilized by each perturbation condition and each cell at varying degrees via the attention mechanism. The incorporation of dot-product cross-attention into the linearly-decoded variational autoencoder gives the rise to the explanatory power of CellCap. A graphical illustration of CellCap model is shown below:
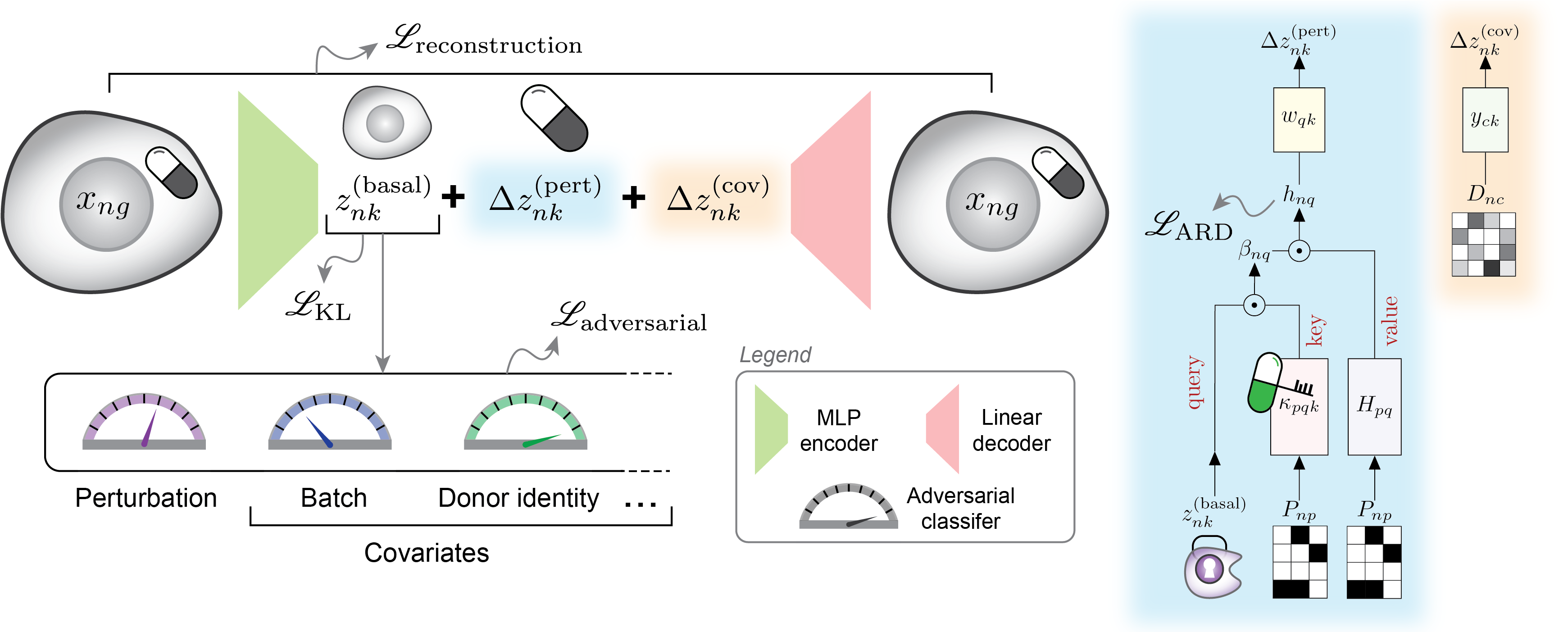
CellCap’s primary objective is to model the relationship between cell state and perturbation response. It achieves this by employing a generative model that is both simple and flexible, utilizing learnable parameters and latent variables with clear interpretations. These parameters are detailed below:
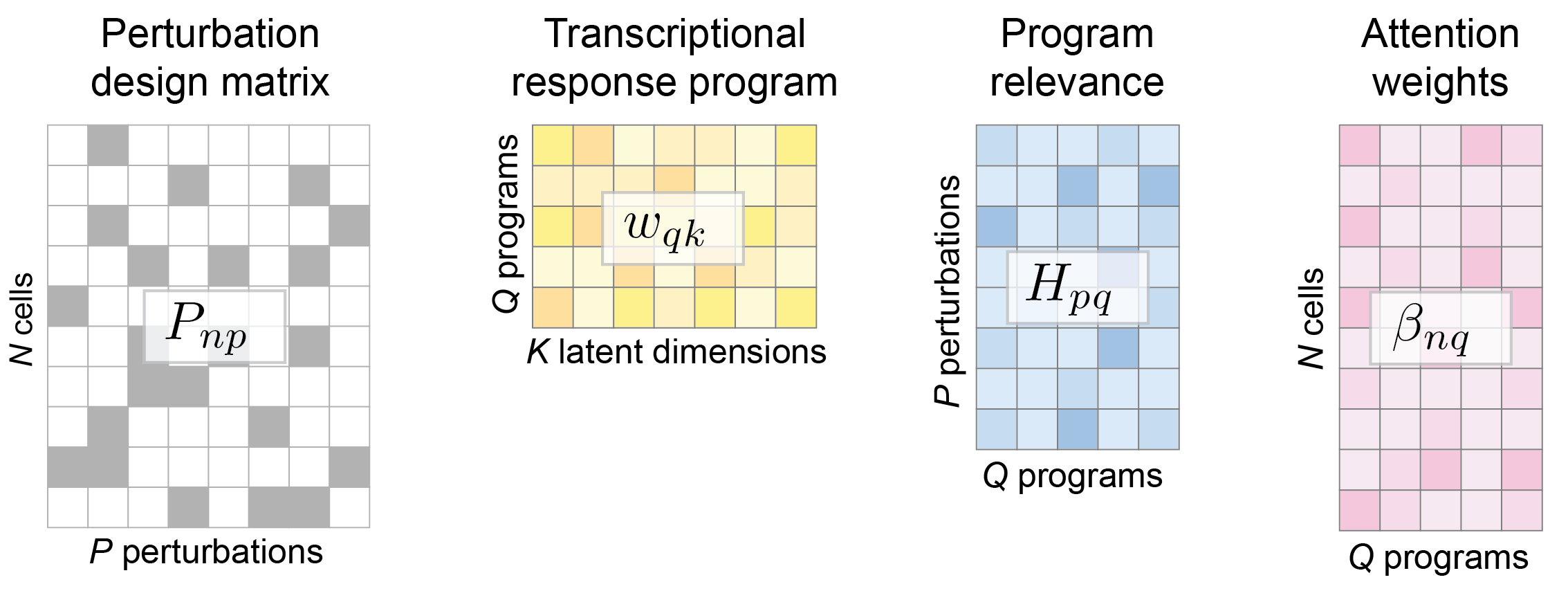
Perturbation design matrix P: The experimental design is represented as a standard design matrix, which specifies the perturbation condition for each cell. A value of 1 within the design matrix P indicates that a cell has received a perturbation, and each cell can have multiple non-zero values to account for combinatorial perturbations.
Dictionary of transcriptional programs w: Each transcriptional program is signified by a row of w and has the same dimension as basal state z. These learnable programs denote transcriptional activation or deactivation of sets of genes, and are used by each perturbation and each cell state by varying degrees.
Program relevance H: A weight matrix that captures the relevance of a transcriptional program to a perturbation condition. CellCap uses the Bayesian automatic relevance determination mechanism to keep this matrix sparse and interpretable.
Attention weights β: CellCaps matches the basal state z of each cell to each transcriptional programs via a learnable set of perturbation keys κ using the dot-product attention mechanism.
Is CellCap suitable for modeling your dataset?
Before you proceed to run CellCap on your dataset, it is important to consider if CellCap’s modeling assumptions faithfully represent your case. The most important assumptions are as follows:
(a) The experiment contains a relevant control group (i.e. unperturbed cells, non-targeting guides in CRISPR screens), and all possible cell states are well-represented in the control group.
(b) Perturbations effects are generally weak, in the sense that cells largely retain their original identity and can be smoothly connected to control cells. Strong perturbations, cell passaging, and long post-perturbation observation intervals may result in significant remodeling of cell identities and obscure their connection to basal states.
What biological questions does CellCap aim to address?
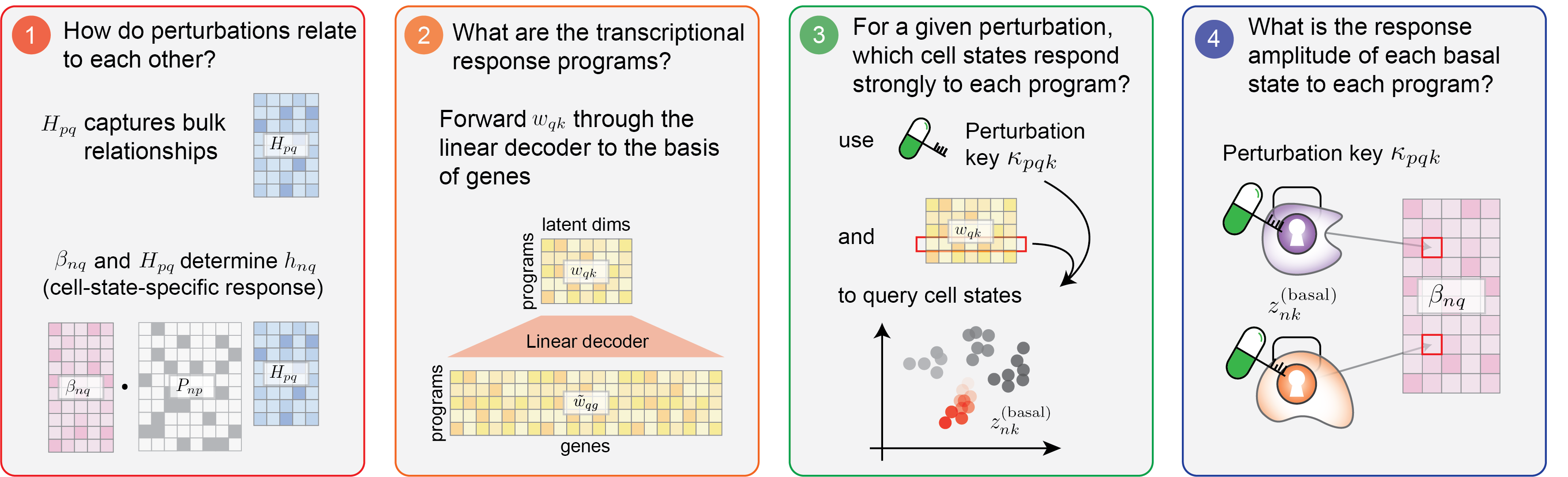
CellCap’s design affords it high interpretability, empowering researchers to understand cellular mechanisms after perturbation across a range of downstream biological investigations. In particular, understanding the relationship between cell state and perturbation response would facilitate addressing multiple biological questions. We list a few suggested questions here, and users can utilize the explainable components of CellCap mentioned above to answer them:
Where to go next?
For additional technical details, use cases, and comprehensive evaluation of CellCap, please refer to the associated preprint. The open-source code can be access from the project GitHub repository. The best place to start your journey is by referring to CellCap’s documentation.