What is Cellarium Cell Annotation Service (CAS)?
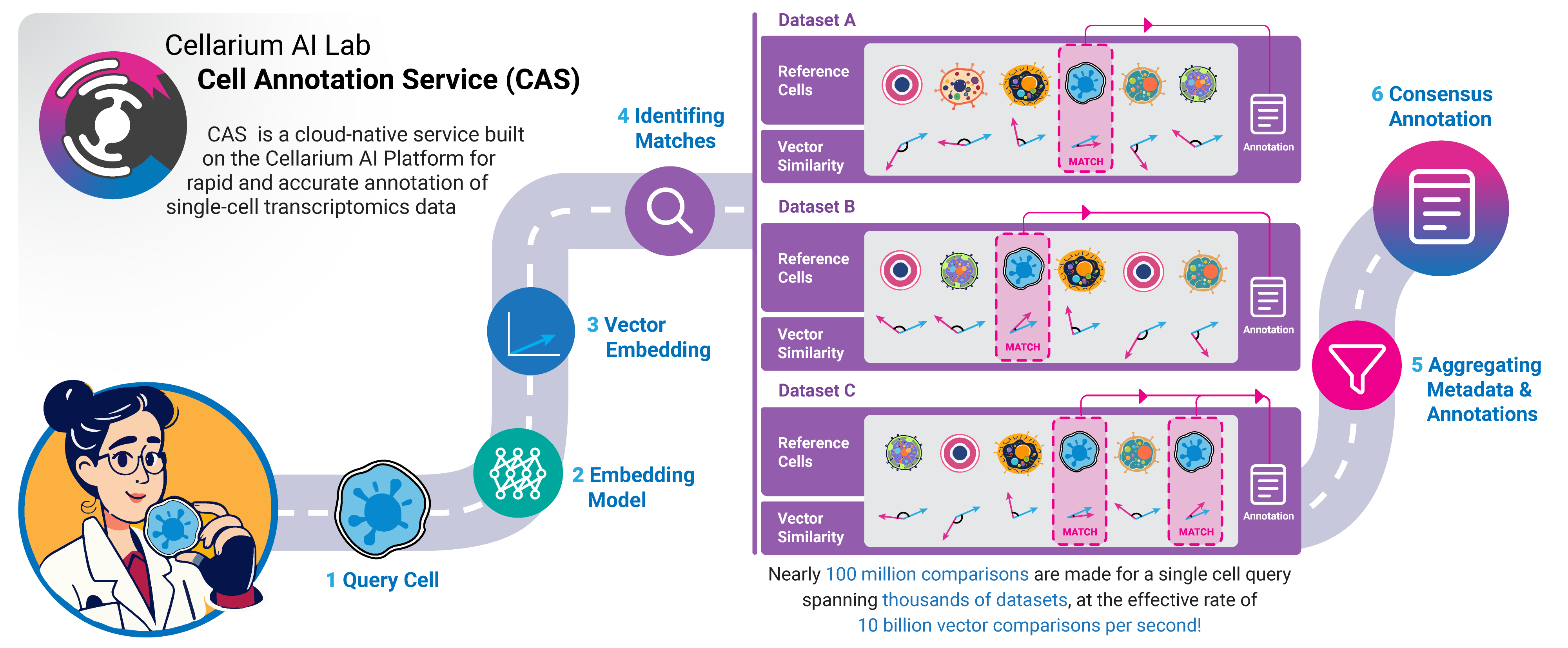
The Cell Annotation Service (CAS) is a cloud-native tool built on the Cellarium AI Platform for rapid and accurate annotation of single-cell transcriptomics data. At its core, CAS functions as an inverse search engine for single-cell data, leveraging compact, information-dense vector embeddings and an approximate nearest neighbor search engine to quickly identify transcriptionally similar cells from a vast reference repository. The reference data repository currently holds nearly 100 million single cells, enriched with experimental metadata and informative labels. By integrating metadata from similar cells, CAS generates high-quality annotations for new datasets.
One of the main challenges in single-cell analysis is identifying cell types and states in new datasets—a task that traditionally requires clustering, differential analysis, and manual marker gene identification based on literature. CAS automates and accelerates this process by providing a community-consensus set of annotations for each cell. These annotations can either serve as final results or as a reliable starting point for further refinement.
Similar to how search engines index and make internet data discoverable, CAS organizes single-cell data to facilitate biological discovery. It plays a critical role in understanding disease mechanisms by identifying similarities and differences across cellular measurements in diverse contexts. By matching new data with similar reference cells, CAS offers seamless, context-aware annotations that streamline complex analyses.
How does CAS work?
CAS operates by building a vector search index derived from low-dimensional embeddings of a comprehensive repository of publicly available single-cell transcriptomics data. Currently, the CAS reference dataset comprises the entire Chan Zuckerberg CELLxGENE Discover data catalog. This reference database includes rich metadata such as cell type, disease state, tissue origin, development stage, and more. In future iterations, the reference catalog will be expanded to incorporate additional datasets. When a user queries CAS with their single-cell transcriptomics data, the system maps the input data into the same vector space as the reference dataset. It then performs an approximate nearest neighbor search to identify similar cells within the reference database. Based on these similar reference cells, CAS generates label summary statistics, providing comprehensive annotations for each query cell. The methodology employed by CAS is comparable to how inverse search engines (such as image search techniques) index and retrieve information from vast amounts of internet data, making it accessible and interpretable. The embeddings used by CAS are generated using distributed machine learning models implemented in the Cellarium ML Library.
What are the current and upcoming features of CAS?
Currently, CAS offers cell type annotation as its primary API. In the coming weeks and months, additional functionalities will be incrementally introduced, including:
- Returning various CAS internal cell embeddings along with annotations
- Providing summary statistics beyond cell type annotation, such as disease and developmental stage
- Identifying published datasets containing cells similar to the queried ones
How can I try CAS?
Please join us today and explore CAS during our public beta phase by signing up below. Shortly after you sign up, we will contact you with your API key and detailed instructions about the next steps. The initial offering is for beta testing, with a weekly and total usage quota sufficient for evaluation purposes. Your feedback will help enhance the service and better support the scientific community. As a token of appreciation, each time you provide feedback on an annotated dataset, your account will be credited with additional quota. Enterprise users needing higher quotas are encouraged to contact us directly.
CAS Python Client Library
The easiest way to interface with the CAS API is through the CAS Python Client Library. We will send you detailed instructions regarding installing the client library, along with a quickstart tutorial. The links to those resources are provided below for your reference.
Acknowledgements
CAS was co-developed by the members of the Cellarium AI Lab at Broad Institute’s Data Sciences Platform and 10x Genomics, with additional support from BICAN Center for Human Brain Variation. Currently, the embedding model powering CAS is the same as the cell annotation pipeline offered by 10x Genomics. Future models and features may evolve separately in alignment with the development roadmaps of each organization.
CAS Terms of Service
You will be asked to read and accept CAS terms of service and privacy policy during the sign up. Please note that usage of CAS is limited by our acceptable use policy. Besides the feedback that you may electively provide to us, we are not intentionally storing any data you submit to CAS. While we take data and application security very seriously, CAS is not currently certified to operate within federal security compliance certifications such as FISMA or FedRAMP. Therefore, we advise you against submitting sensitive and/or identifiable data to CAS during the current beta testing phase.