What is CellBender?
CellBender is a software package designed to remove technical artifacts from high-throughput single-cell omics data, including scRNA-seq, snRNA-seq, and CITE-seq. A common application of CellBender is to mitigate ambient RNA contamination, as illustrated in the schematic below. The schematic depicts a background of “ambient” molecules from the cell suspension media (black debris) being encapsulated into droplets alongside individual cells (red) and barcoded beads (green hexagons):
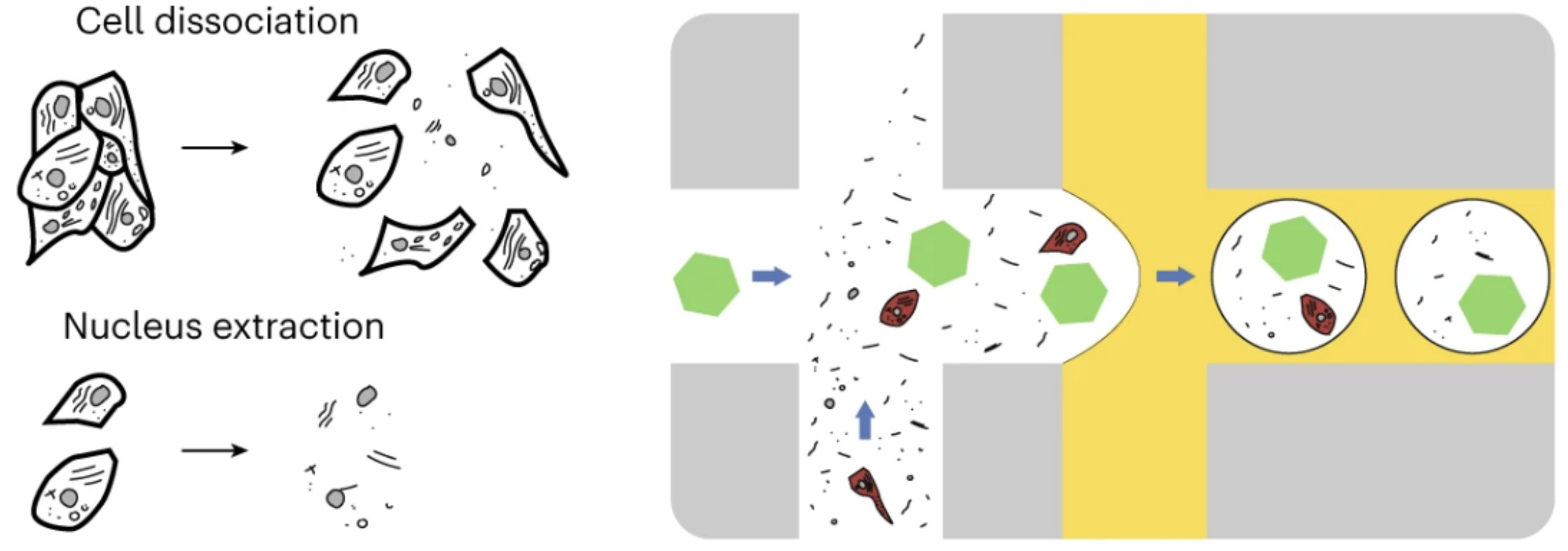
(left) Cellular dissociation and generation of cell suspension prior to single-cell sequencing; (right) Background “ambient” molecules are packaged up with each cell (or cell nucleus).
By correcting technical artifacts, CellBender aims to improve the accuracy and reliability of downstream analyses. This can lead to more accurate identification of cell types, better detection of gene expression patterns, removal of confounding background noise, and enhanced understanding of biological processes at the single-cell level. CellBender operates end-to-end starting from the raw counts, is fully unsupervised, is agnostic to the nature of the measured molecular feature (for example, mRNA, protein and so on) and requires no assumptions or prior biological knowledge of either cell types or cell type-specific gene expression profiles.
Overall, CellBender helps researchers to obtain clearer and more accurate insights from their scRNA-seq data, ultimately advancing our understanding of cellular heterogeneity, gene regulation, and other biological phenomena.
How does CellBender work?
CellBender simultaneously learns the profile of background noise and the biological signal via a structured deep generative model that incorporate key aspects of the data generation process. A major challenge in distinguishing background noise counts from biological counts for single droplets is the extreme sparsity of counts, such that, without a strong informative prior, the counts obtained from a single droplet do not provide sufficient statistical power to allow inference of background contamination. CellBender uses a neural network to learn the distribution of the underlying biological signal (e.g. gene expression) across all droplets. The learned distribution acts as a prior over cell-endogenous counts, provides a mechanism to share statistical power between similar cells and ultimately improves the estimation of background noise counts. Learning this neural prior of cell states and estimating the background noise profile is performed simultaneously and self-consistently within a variational inference framework, allowing progressively improved separation of endogenous and background counts during model training.
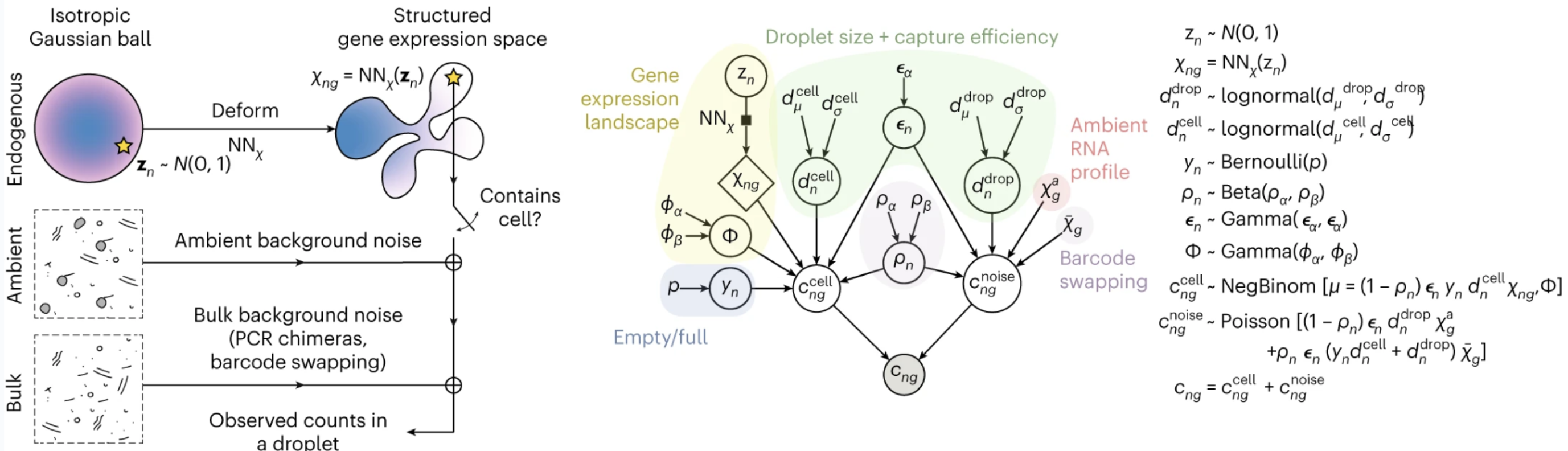
(left) CellBender’s structured deep generative model in a nutshell; (right) The same model shown as a probabilistic graphical model in more detail.
What kind of result should you expect?
CellBender works like a charm especially if your data is severely contaminated, e.g. single-nuclei RNA-seq experiments. The following shows an example from a snRNA-seq dataset generated from human cardiac tissue. We are showing the gene CTNNA3, a protein-coding gene involved in cell-cell adhesion with known and specific expression pattern before (left) and after (right) CellBender. For reference, the two cell clusters with high expression of CTNNA3 after CellBender processing are cardiomyocytes (the middle left cluster) and vascular smooth muscle cell (the top right cluster).
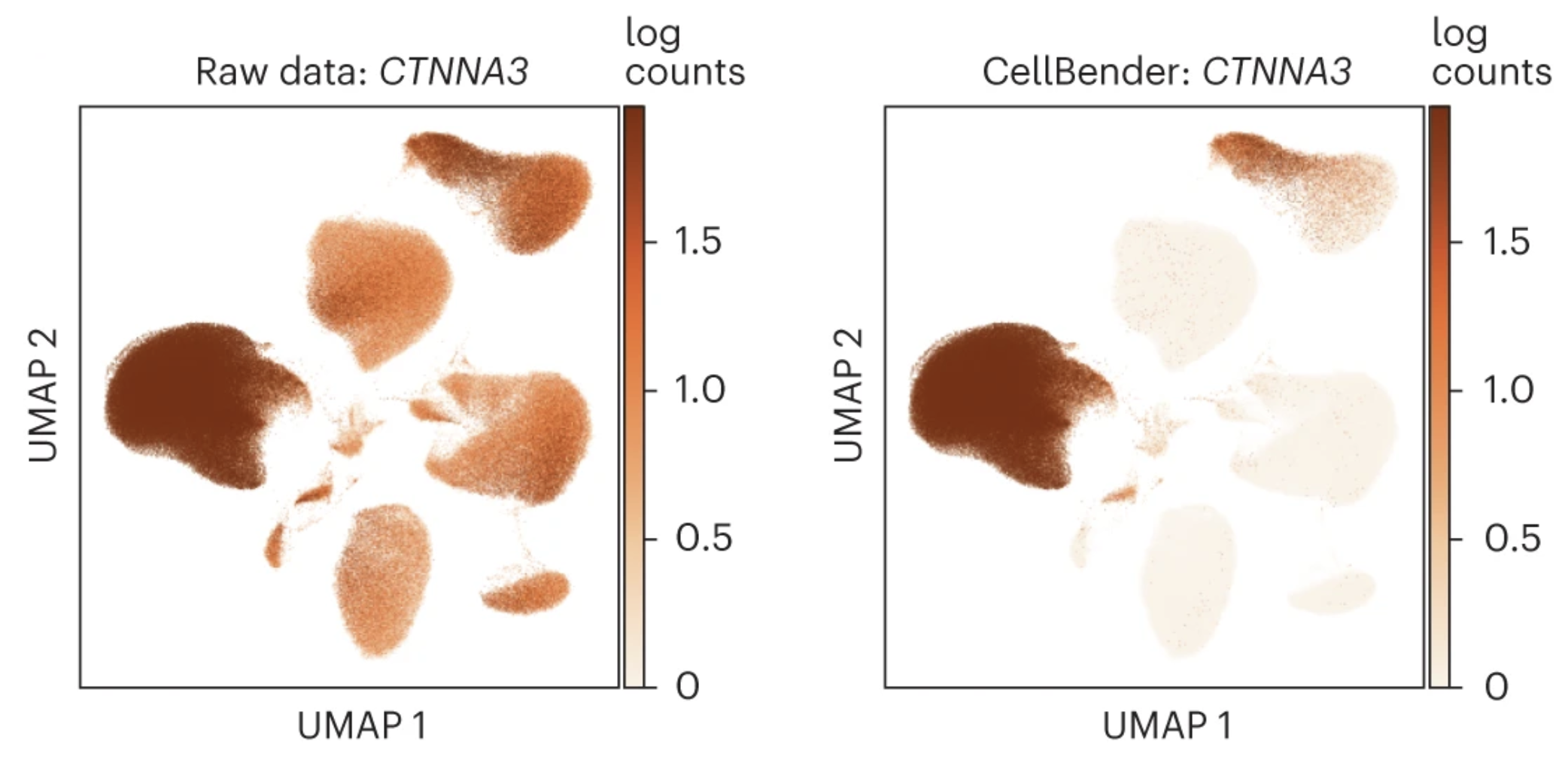
UMAP plots show counts of CTNNA3, a protein-coding gene involved in cell–cell adhesion with known cell type expression, before and after CellBender noise removal. Gene expression assay results become much more cell-type specific.
Where to go next?
For additional technical details, use cases, and comprehensive evaluation of CellBender, please refer to the associated publication. The open-source code can be accessed from the project GitHub repository. The best place to start your journey is by referring to CellBender’s documentation.