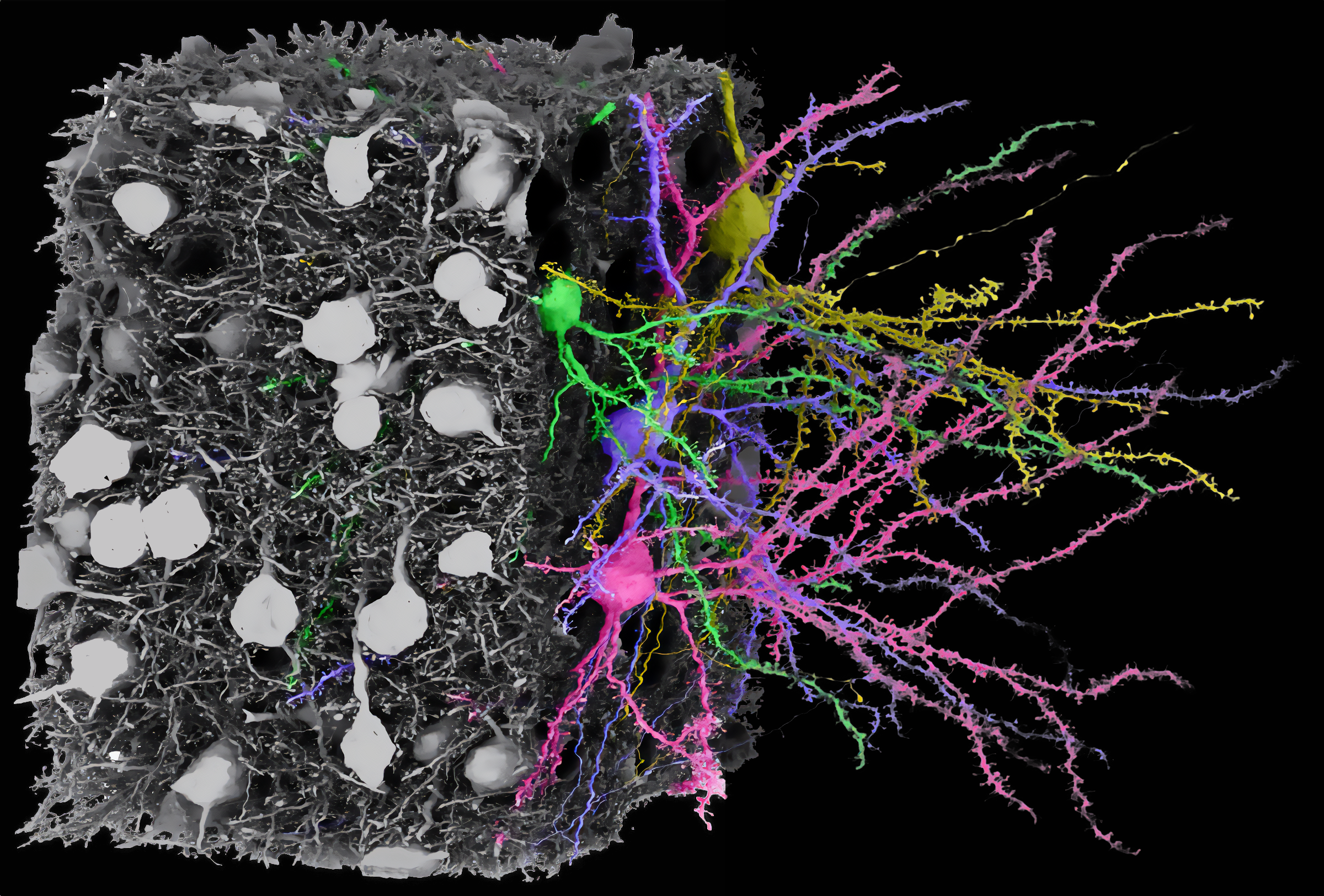
What is SynapseCLR?
SynapseCLR addresses a major bottleneck in electron microscopy (EM) connectomics, a field aiming to describe complete cellular connectivity patterns in nervous tissue using 3D EM reconstructions of ex vivo tissue samples. 3D EM resolves all cells and organelles, informing about nervous tissue
composition and function. Insights from 3D EM could aid efforts to identify neurological disease therapies or design better artificial intelligence; however, analysis is currently limited, with often-manual proofreading, exploration, and annotation of individual structures (e.g., synapses) taking years. SynapseCLR aims to automate 3D EM analysis. By producing compact and information-rich vector representations of segmented structures, SynapseCLR accelerates proofreading (by separating defective versus valid segmentations), annotation (with imputation using few manual annotations), and exploration (with dataset-wide querying of segmented objects by structural similarity). As the name suggests, SynapseCLR is tailored to the study of chemical synapses. However, the underlying principle and the implementation can be readily repurposed to studying arbitrary 3D EM segmented objects.
How does SynapseCLR work?
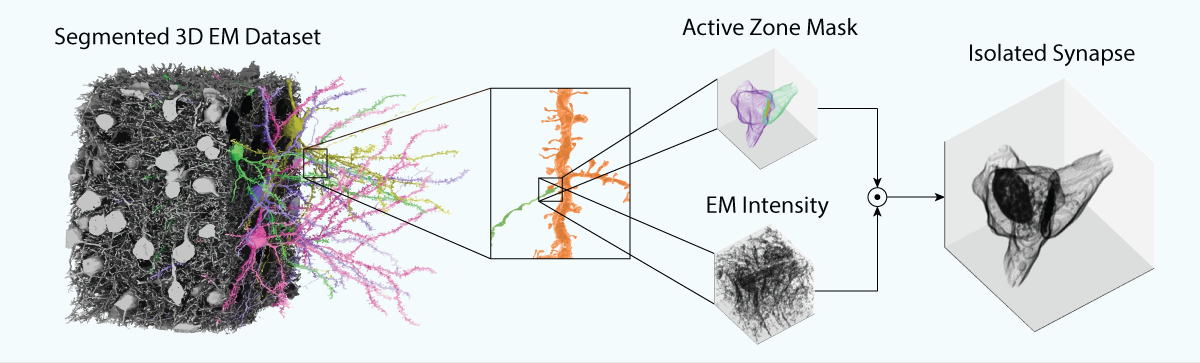
SynapseCLR takes as input a collection of segmented 3D EM volumetric image chunks, that is, fixed-sized crops of a 3D EM dataset along with precomputed segmentation masks delineating the pre-synaptic process, post-synaptic process, and the synaptic cleft region. The segmentation masks are used to isolate the 3D EM morphology of individual synapses from the dense neuropil:
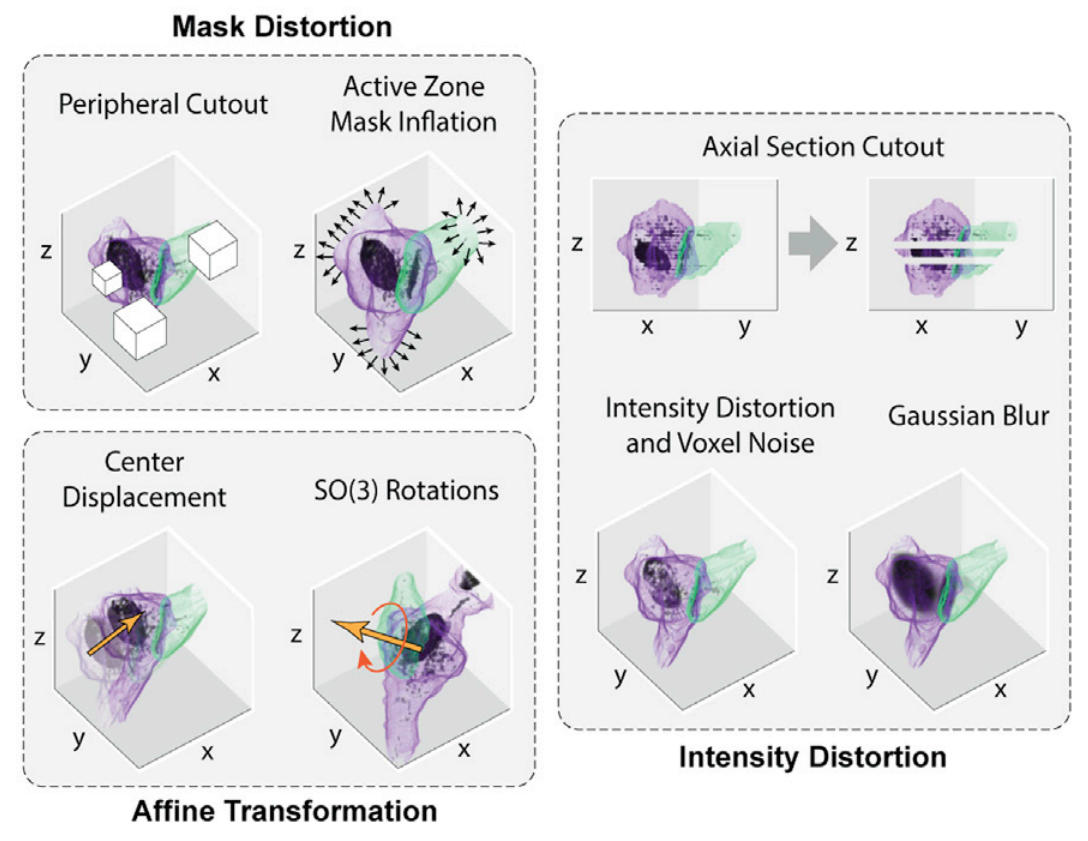
SynapseCLR proceed to learn robust low-dimensional embeddings (“representations”) of chemical synapses using the principle of contrastive learning. In brief, isolated synapses are subjected to a collection of stochastic technical augmentation transformations designed to preserve the biological identity of synapses while distilling out the information related to imaging artifacts, pose, and location. The augmentation include various mask distortions, isometric affine transformations, and intensity distortions:
We use a 3D-ResNet18 CNN as the base representation learning model (“backbone”) and train it using the SimCLR methodology in conjunction with the mentioned augmentation transformation mentioned:
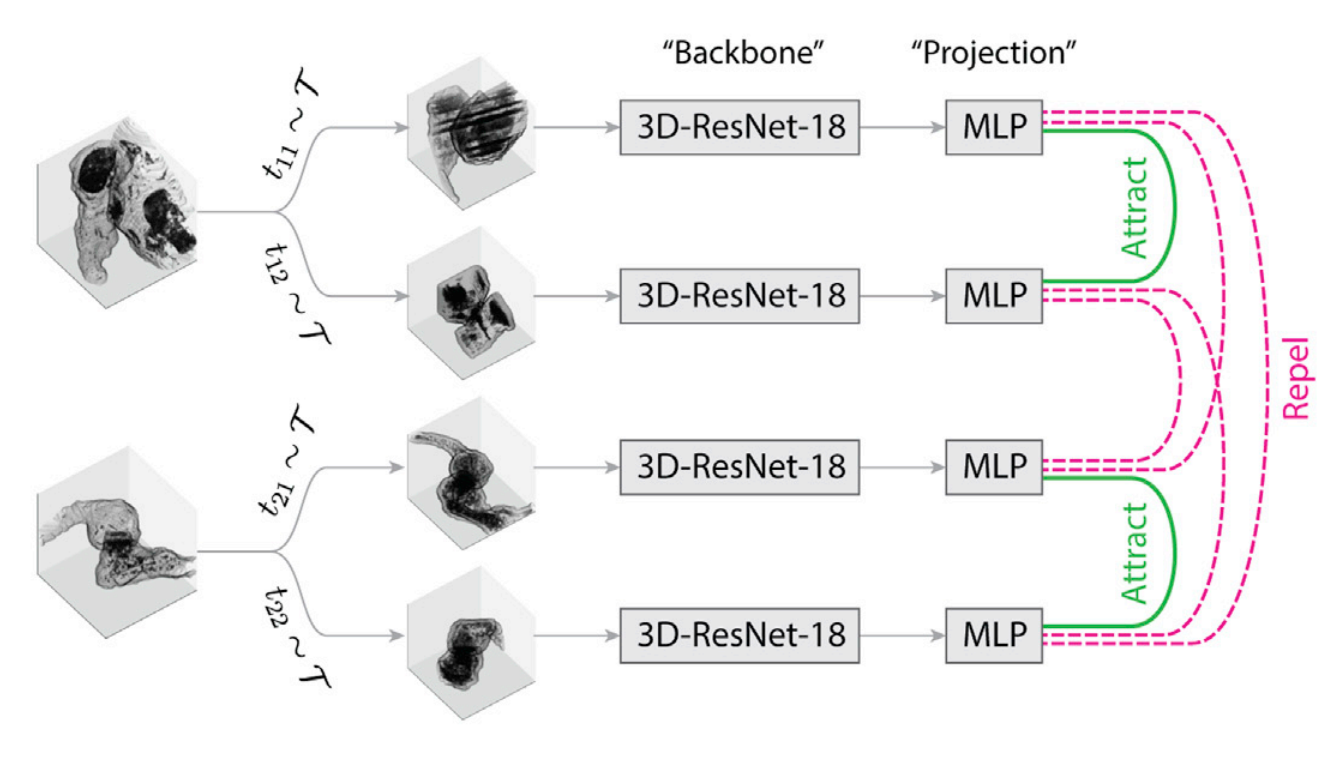
What biological problems SynapseCLR solve?
The representations generated by SynapseCLR can be used to streamline various connections tasks. These include few-shot probabilistic imputation of annotations, dataset-wide query of synapses based on morphological similarity, one-shot outlier detection, accurate cell-type annotation, and charting out a data-driven manifold of synaptic morphological variation:
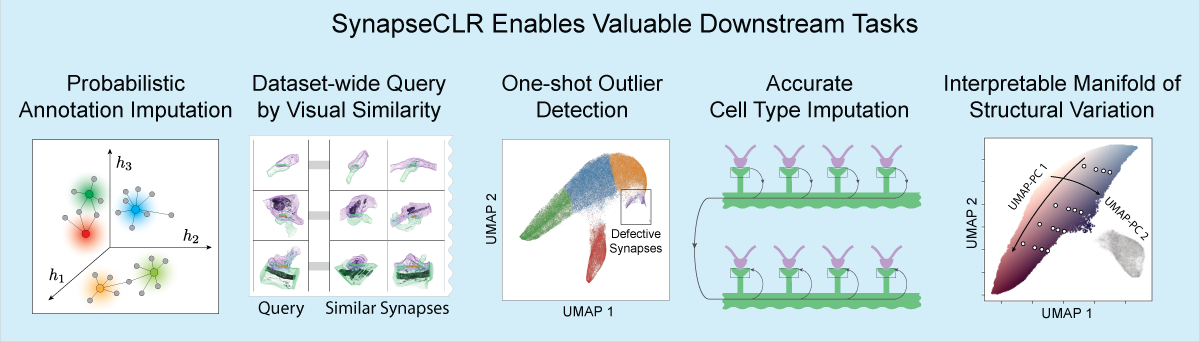
Where to go next?
For additional technical details, use cases, and comprehensive evaluation of SynapseCLR, please refer to the associated publication. The open-source code, pretrained models, and data can be access from the project GitHub repository.