What is voltage imaging?
Voltage imaging, like other forms of fluorescence imaging, uses a fluorescent indicator to report electrophysiological activity. Fluorescent voltage indicators are diverse, including voltage sensitive dyes (such as BeRST [1]) and genetically-encoded voltage indicators (such as QuasAr [2]). Together with genetically encoded light gated cation channels (e.g. Channelrhodopsins-2 [3]), researchers can optically manipulate and record a large field of neurons in parallel. The following figure schematically depicts a typical voltage imaging setup:
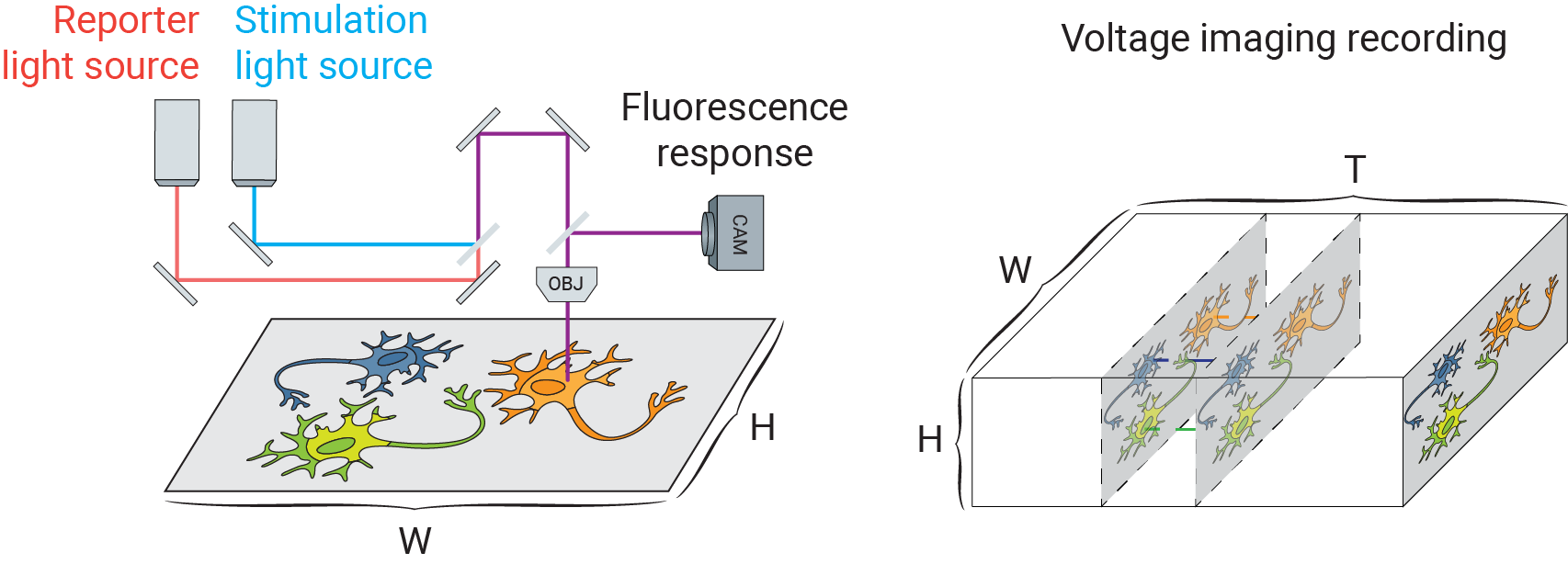
The true potential of this powerful toolbox, however, is often constrained by a low signal-to-noise ratio (SNR) due to shot noise and camera noise, limiting their application in reconstructing small-scale electrophysiology (EP) features such as detecting subthreshold events.
What is CellMincer?
CellMincer is a machine learning tool for self-supervised denoising of voltage imaging datasets. Importantly, CellMincer does not require clean data for model training. Instead, it operates on the principle of self-supervision to tease out the signal component from noise, circumventing the need for clean training data altogether. The movie below demonstrates typical results obtained using CellMincer. The raw voltage imaging recording and temporal activity traces from several pixels is shown on the left. The same data is shown on the right following CellMincer self-supervised denoising:
How does CellMincer work?
CellMincer operates on the principle of self-supervision, and is methodologically a generalization of the powerful Noise2Self blind denoising algorithm [4] to spatiotemporal data. The Noise2Self technique can be described as follows. Suppose a sparse set of pixels are masked out from a noisy image, and a neural network is trained to predict the value of the sparsely masked pixels from the rest of the image, i.e. the majority of pixels. Assuming that the noise in the masked pixels is uncorrelated with the rest of the pixels, the optimal predictor can at best predict the noiseless signal component; in practice, it can excel at this task given the strong spatial correlations and redundancy in biological images. It follows that the optimal masked pixel predictor in turn behaves as an optimal pixel denoiser. Noisy data itself provides the needed evidence for teasing out the signal component, circumventing the need for clean training data.
One of the main challenges in extending self-supervised image denoising approaches to spatiotemporal data, such as voltage imaging recordings, is that these datasets contain thousands of frames, and that each frame is individually too signal-deficient to self-supervise its own denoising. At the same time, GPU hardware memory constraints and efficient training considerations prevent us from ingesting and processing entire voltage imaging movies with neural networks to exploit frame-to-frame correlations. The middle ground strategy adopted by several authors is to process the movie in overlapping and truncated local denoising temporal contexts, i.e. chunks of adjacent frames. In order to obtain satisfactory results, however, one needs to operate on very large temporal windows— much like the role of larger context sizes in improving the quality of large language models. We solve this problem by first precomputing a large number of spatiotemporal spatiotemporal auto-correlations at multiple length scales from the entire movie:
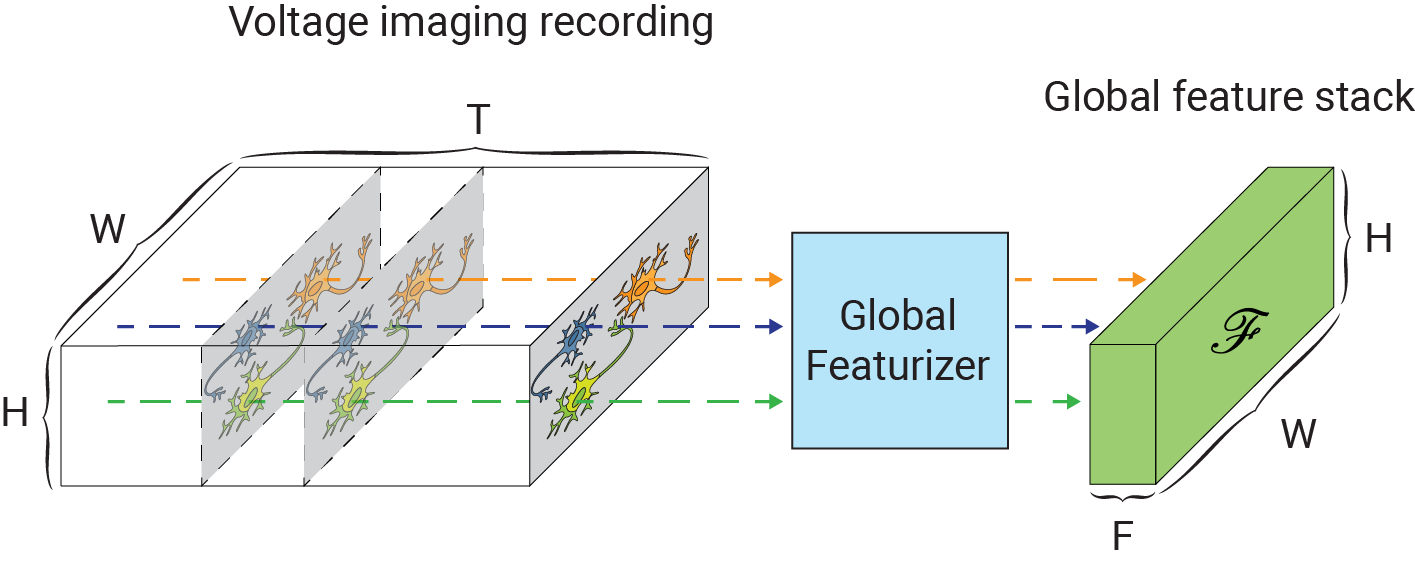
These precomputed global features are used as a conditioner for the local denoiser neural network, effectively augmenting the local context with long-range information. CellMincer uses a two-stage spatiotemporal data processing deep neural network architecture for local denoising, comprising a frame-wise 2D conditional U-Net module for spatial feature extraction, followed by a pixelwise 1D convolutional module for temporal data post-processing. The model architecture and self-supervised training procedure of CellMincer’s denoiser network are shown below:
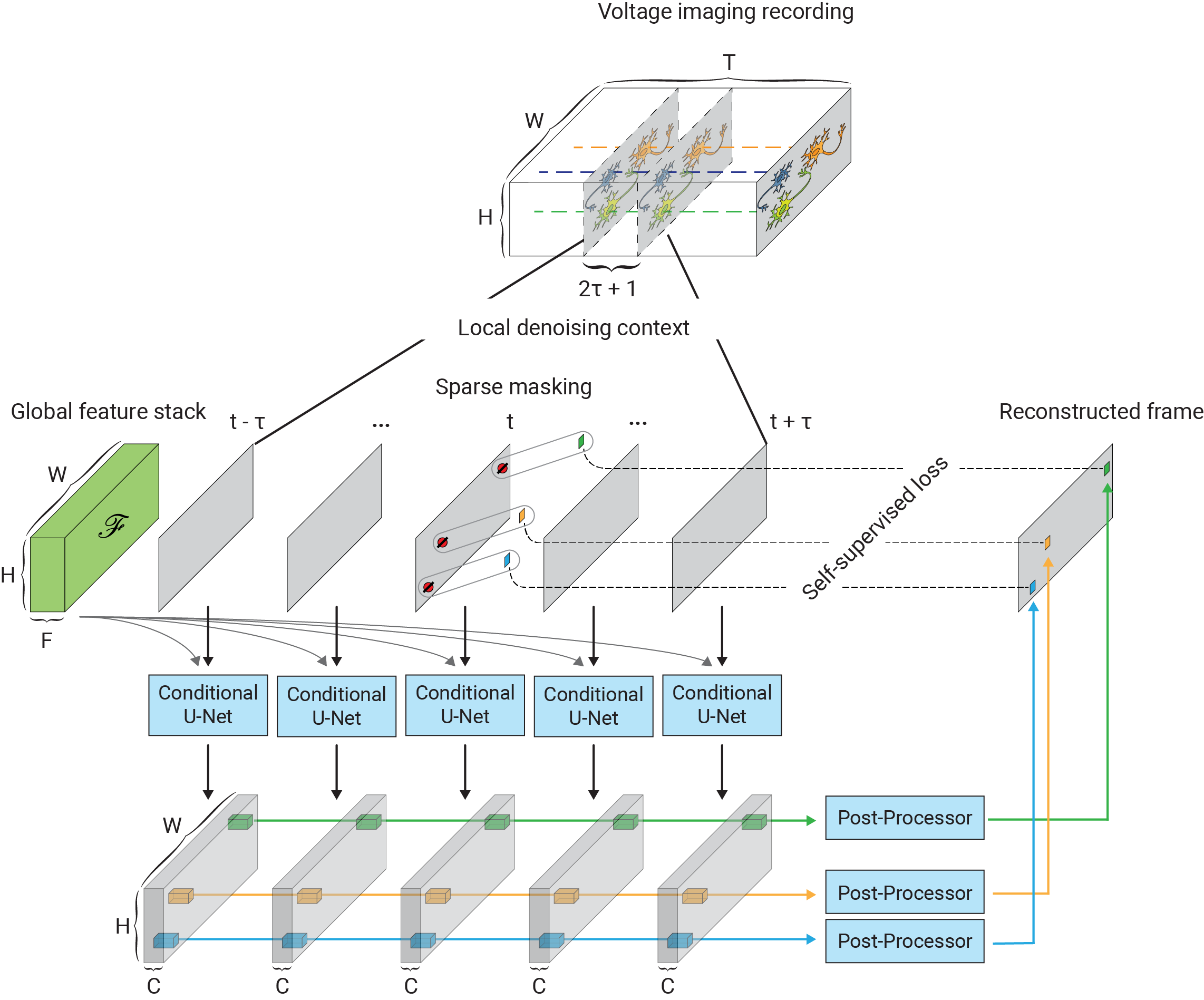
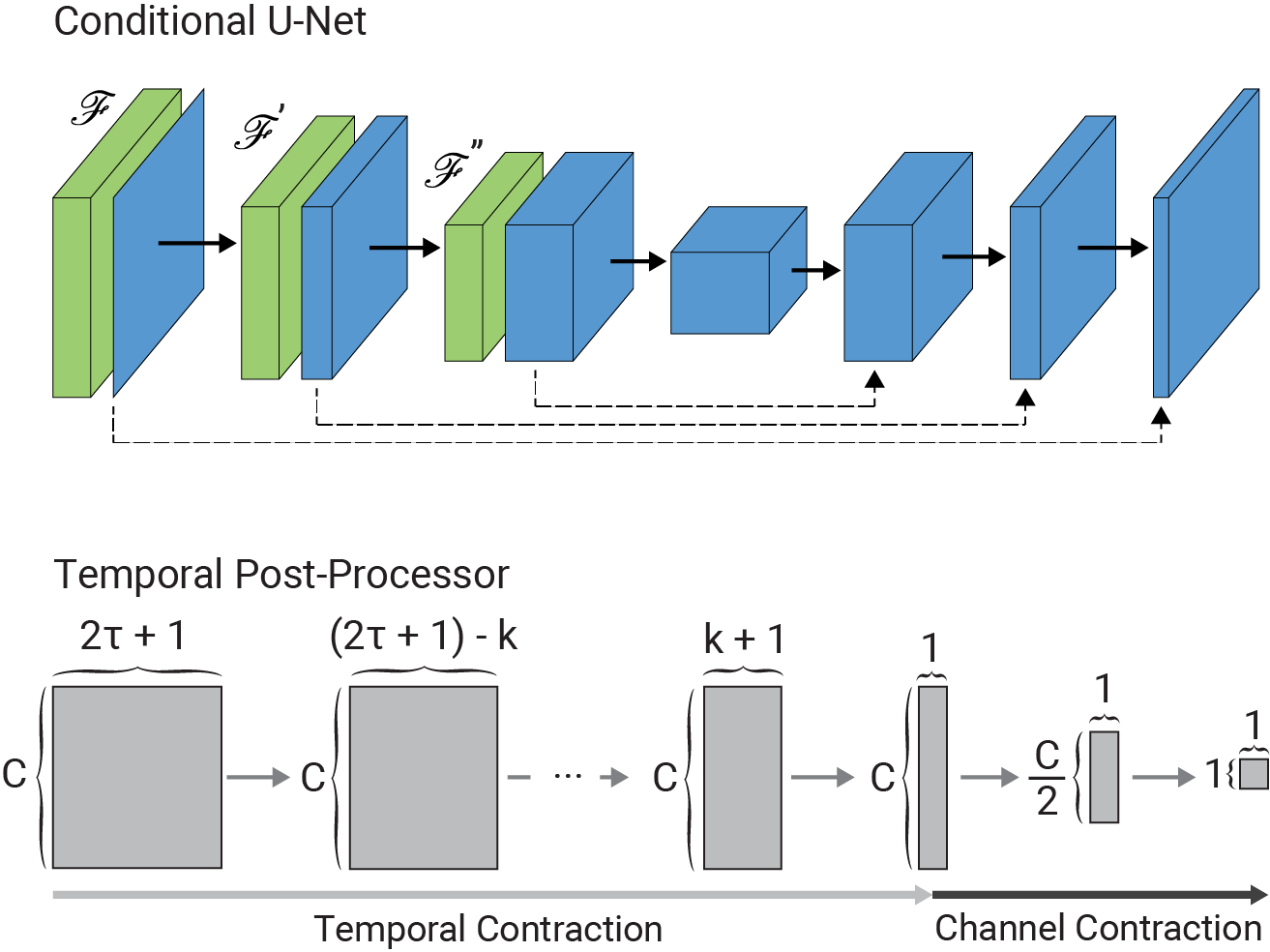
The architecture of the conditional U-Net modules and the temporal post-processor is as follows:
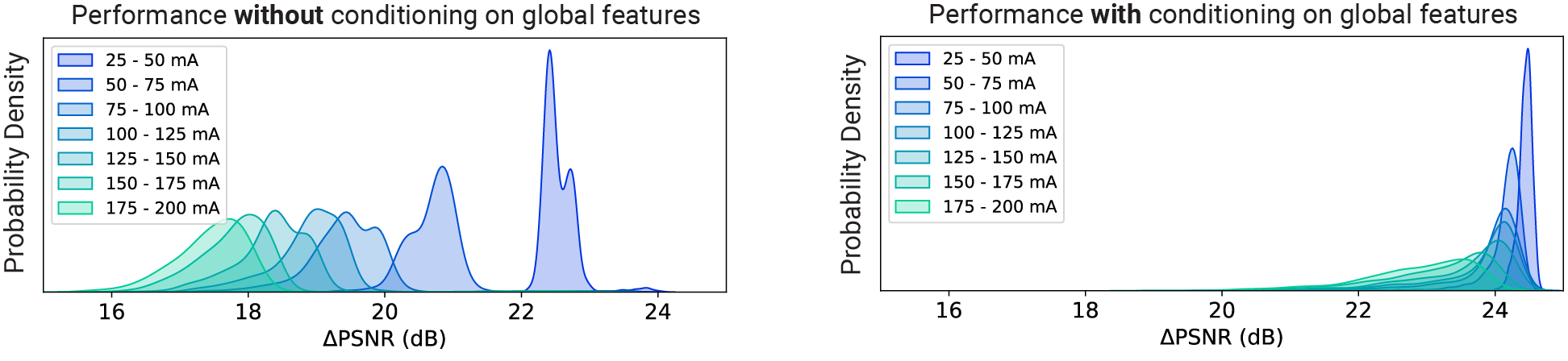
How much does conditioning improve the model’s performance?
A lot. Below, we are showing the frame-wise peak signal-to-noise ratio (PSNR) gains achieved by CellMincer on a synthetic voltage imaging dataset with (right) and without (left) conditioning on precomputed global features. We notice that not only conditioning results in a more uniform operation across different experiments, but also this technique results in approximately 5 dB additional gain in PSNR, i.e. 3-fold further reduction in noise on average:
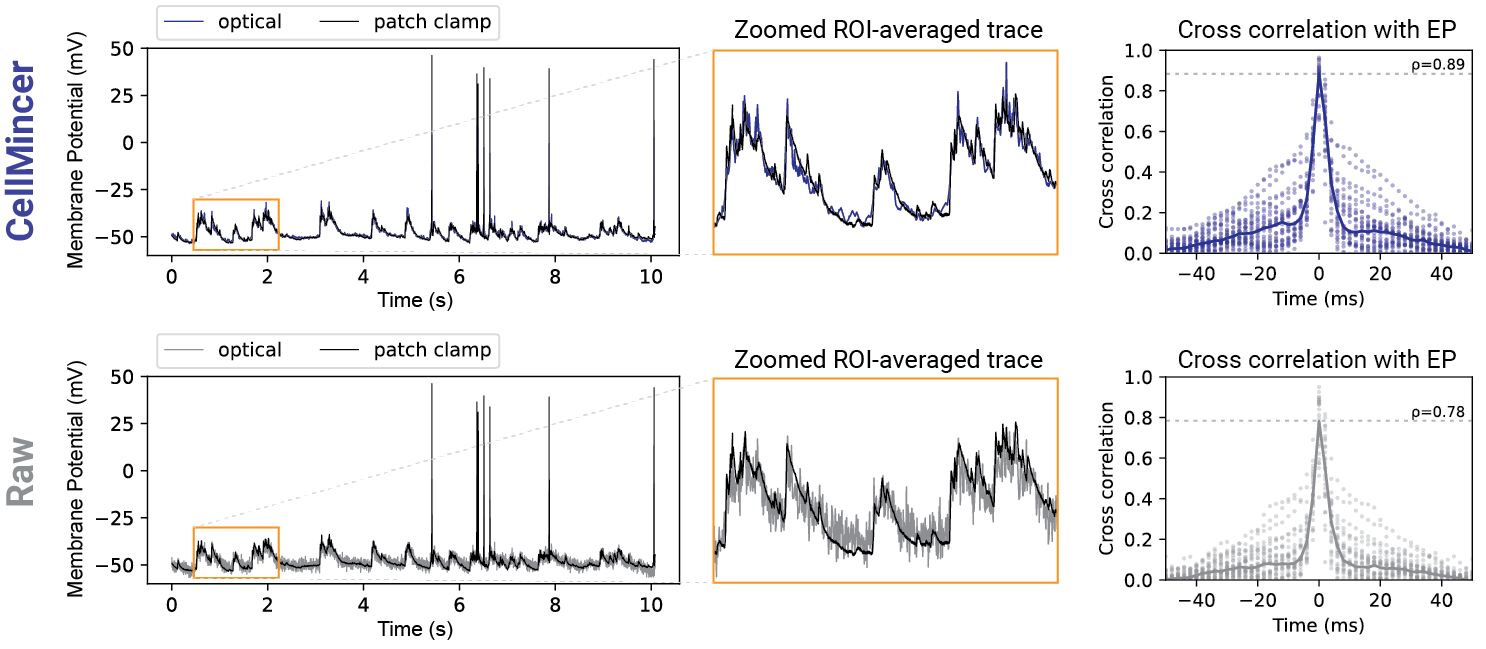
How well does CellMincer work in practice?
In our benchmarking experiments with both synthetic and real data with paired electrophysiology (EP) recordings, we found an average PSNR gain of 24 dB compared to the raw data (an increase of 2 dB over the next best benchmarked method), a 14 dB reduction in high-frequency (>100 Hz) noise (a further reduction of 10.5 dB from the next best method), a 5-10 percentage point increase of F1-score in detecting sub-threshold events compared to the other algorithms and across all voltage magnitudes in the 1-10 mV range (in which the baseline F1-score ranges from 5-14%), and more than 20% increase in the cross-correlation between low-noise EP recordings and voltage imaging. A comparison of voltage imaging data overlaid on EP recordings (top: CellMincer-denoised, bottom: raw) is shown below. Please refer to the associated preprint for further benchmarking with existing methods, as well as a case study in which we quantify the degree to which CellMincer increases the statistical power to differentiate subtle functional phenotypes in a typical case-control hypothesis testing scenario.
Where to go next?
For additional technical details, use cases, and comprehensive evaluation of CellMincer, please refer to the associated preprint. The open-source code, pretrained models, and data can be access from the project GitHub repository.
References
[1] Huang, Yi-Lin, Alison S. Walker, and Evan W. Miller. “A photostable silicon rhodamine platform for optical voltage sensing.” Journal of the American Chemical Society 137.33 (2015): 10767-10776.
[2] Hochbaum, Daniel R., et al. “All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins.” Nature Methods 11.8 (2014): 825-833.
[3] Nagel, Georg, et al. “Channelrhodopsin-2, a directly light-gated cation-selective membrane channel.” Proceedings of the National Academy of Sciences 100.24 (2003): 13940-13945.
[4] Batson, Joshua, and Loic Royer. “Noise2self: Blind denoising by self-supervision.” International Conference on Machine Learning. PMLR (2019): 524-533.